Chapter 20 Modelling with XGBoost
We try to predict whether the lines are written by a specific author.
We do Cross Validation using Caret package.Lastly we wish to examine the feature importance of the variables. This is shown in the flipped bar chart.
We then use the model to predict the authors.
makeFeatures <- function(train) {
labeledTerms = makeDTM(train)
## Preparing the features for the XGBoost Model
features <- colnames(labeledTerms)
for (f in features) {
if ((class(labeledTerms[[f]])=="factor") || (class(labeledTerms[[f]])=="character")) {
levels <- unique(labeledTerms[[f]])
labeledTerms[[f]] <- as.numeric(factor(labeledTerms[[f]], levels=levels))
}
}
return(labeledTerms)
}
labeledTerms = makeFeatures(train)
labeledTermsTest = makeFeatures(test)
colnamesSame = intersect(colnames(labeledTerms),colnames(labeledTermsTest))
labeledTerms = labeledTerms[ , (colnames(labeledTerms) %in% colnamesSame)]
labeledTermsTest = labeledTermsTest[ , (colnames(labeledTermsTest) %in% colnamesSame)]20.1 Add features
We add the following features to the model
Number of words in the line
Sentiment Score per line
labeledTerms$len = train$len
labeledTermsTest$len = test$len
labeledTerms$sentiScore = getSentimentScore(train)
labeledTermsTest$sentiScore = getSentimentScore(test)20.2 Creating the XGBoost Model
labeledTerms$author = as.factor(train$author)
levels(labeledTerms$author) = make.names(unique(labeledTerms$author))
formula = author ~ .
#Please uncomment if you want to do Cross Validation
# fitControl <- trainControl(method="cv",number = 5,classProbs=TRUE, summaryFunction=mnLogLoss)
#
# xgbGrid <- expand.grid(nrounds = 500,
# max_depth = 3,
# eta = .05,
# gamma = 0,
# colsample_bytree = .8,
# min_child_weight = 1,
# subsample = 1)
fitControl <- trainControl(method="none",classProbs=TRUE, summaryFunction=mnLogLoss)
xgbGrid <- expand.grid(nrounds = 500,
max_depth = 3,
eta = .05,
gamma = 0,
colsample_bytree = .8,
min_child_weight = 1,
subsample = 1)
set.seed(13)
AuthorXGB = train(formula, data = labeledTerms,
method = "xgbTree",trControl = fitControl,
tuneGrid = xgbGrid,na.action = na.pass,metric="LogLoss", maximize=FALSE)
importance = varImp(AuthorXGB)
varImportance <- data.frame(Variables = row.names(importance[[1]]),
Importance = round(importance[[1]]$Overall,2))
# Create a rank variable based on importance
rankImportance <- varImportance %>%
mutate(Rank = paste0('#',dense_rank(desc(Importance)))) %>%
head(20)
rankImportancefull = rankImportance
ggplot(rankImportance, aes(x = reorder(Variables, Importance),
y = Importance)) +
geom_bar(stat='identity',colour="white", fill = fillColor) +
geom_text(aes(x = Variables, y = 1, label = Rank),
hjust=0, vjust=.5, size = 4, colour = 'black',
fontface = 'bold') +
labs(x = 'Variables', title = 'Relative Variable Importance') +
coord_flip() +
theme_bw()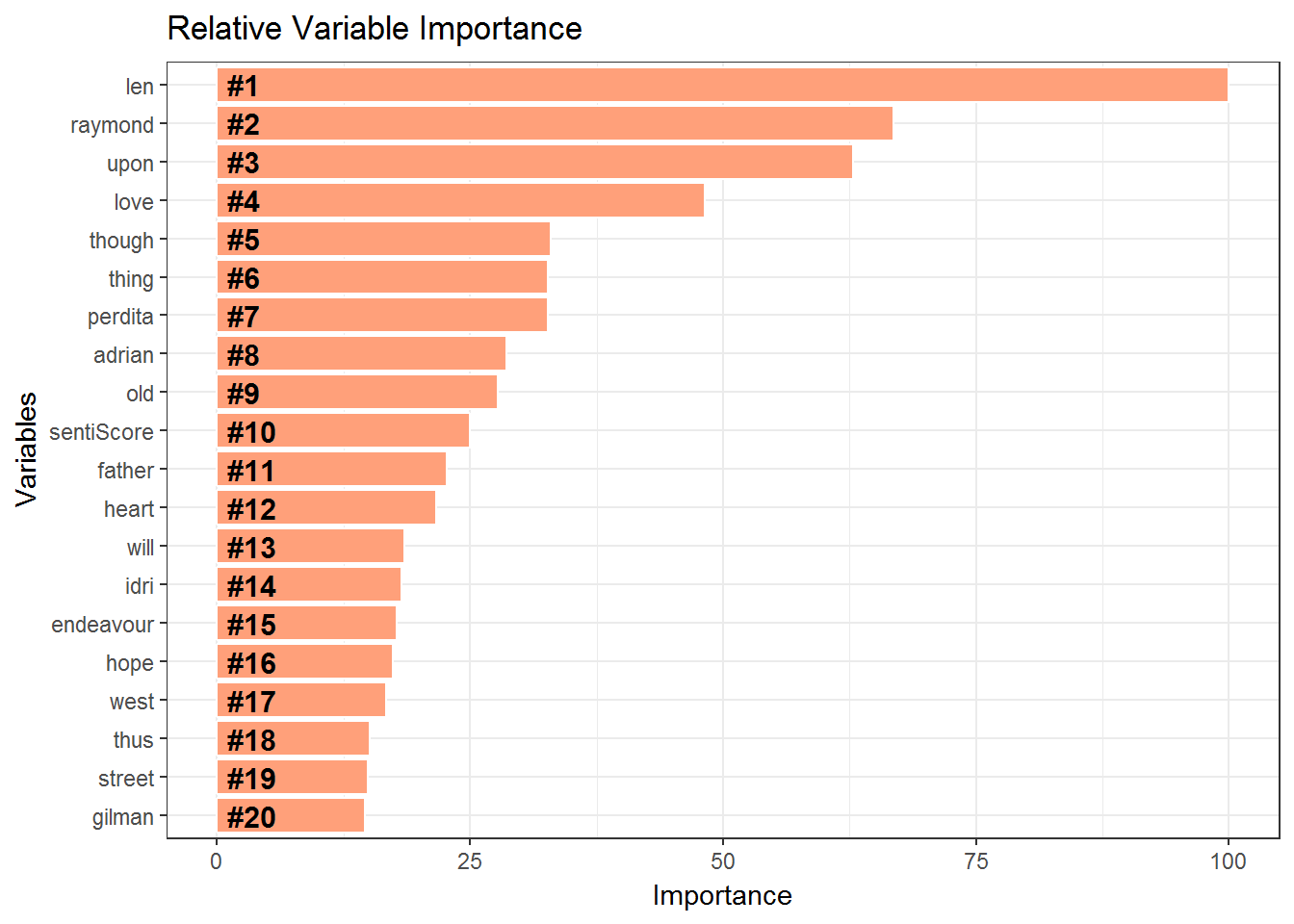
AuthorXGB## eXtreme Gradient Boosting
##
## 19579 samples
## 850 predictor
## 3 classes: 'EAP', 'HPL', 'MWS'
##
## No pre-processing
## Resampling: None